StaR: A Stability-Aware Representation Learning Framework for Spatial Domain Identification
- Topics: spatial transcriptomics, spatial domain identification, representation learning, model robustness
- Skills:
- Programming Languages: Python; PyTorch experience preferred
- Machine Learning: representation learning, clustering, robustness and stability analysis
- Data Analysis: spatial transcriptomics preprocessing and evaluation (ARI, clustering metrics)
- Bioinformatics Knowledge (preferred): familiarity with spatial transcriptomics or scRNA-seq data
- Difficulty: Advanced
- Size: Large (350 hours)
- Mentors: Ziheng Duan (contact person)
Project Idea Description
Spatial domain identification is a fundamental task in spatial transcriptomics (ST), aiming to partition tissue sections into biologically meaningful regions based on spatially resolved gene expression profiles. These spatial domains often correspond to distinct anatomical structures, cellular compositions, or functional microenvironments, and serve as a critical foundation for downstream biological analysis.
Despite rapid methodological progress, most existing spatial domain identification methods are highly sensitive to random initialization. In practice, simply changing the random seed can lead to substantially different clustering results and large performance fluctuations, even when using identical hyperparameters and datasets. This instability severely undermines the reliability, reproducibility, and interpretability of spatial transcriptomics analyses.
Problem: Seed Sensitivity and Unstable Representations
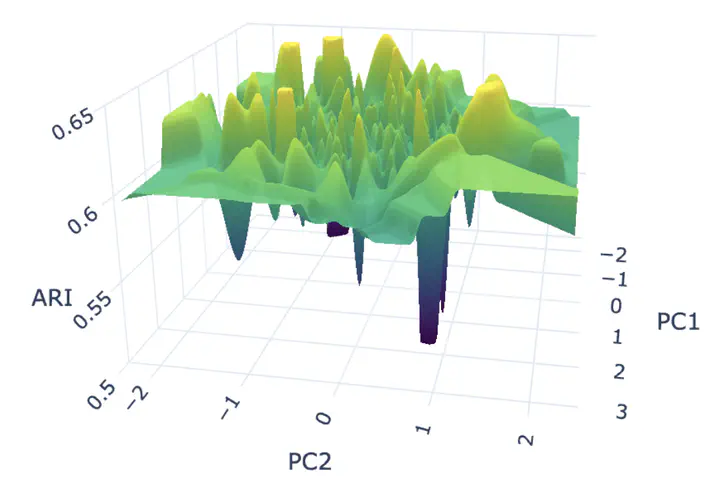
Empirical evidence shows that state-of-the-art spatial domain identification models can exhibit substantial performance variance across random seeds. For example, the Adjusted Rand Index (ARI) may vary from relatively strong performance (e.g., ARI ≈ 0.65) to noticeably degraded yet still reasonable outcomes (e.g., ARI ≈ 0.50) solely due to different random initializations.
By systematically evaluating models across hundreds to thousands of random seeds, we observe that:
- Model performance landscapes are highly rugged, with sharp cliffs and isolated high-performing regions.
- Standard training objectives implicitly favor brittle representations that are not robust to small perturbations in initialization or optimization trajectories.
These observations suggest that instability is not a peripheral issue, but rather a structural limitation of current representation learning approaches for spatial transcriptomics.
Proposed Solution: StaR
This project proposes StaR, a Stability-Aware Representation Learning framework designed to explicitly address seed sensitivity in spatial domain identification.
The core idea of StaR is to learn representations that are robust to perturbations in model parameters and training dynamics, rather than optimizing solely for peak performance under a single random seed. Concretely, StaR introduces controlled noise or perturbations into the training process and encourages consistency across multiple perturbed model instances, guiding the model toward flatter and more stable regions of the parameter space.
By prioritizing stability during representation learning, StaR aims to produce embeddings that:
- Yield consistent spatial domain assignments across random seeds
- Maintain competitive or improved clustering accuracy
- Better reflect underlying biological structure
Project Objectives
- Characterize Instability in Existing Methods
- Systematically quantify seed sensitivity across popular spatial domain identification models.
- Develop Stability-Aware Training Objectives
- Design perturbation-based or consistency-driven losses that encourage robust representations.
- Integrate StaR into Existing Pipelines
- Apply StaR to widely used spatial transcriptomics workflows with minimal architectural changes.
- Evaluation and Benchmarking
- Evaluate StaR using clustering metrics (e.g., ARI) and stability metrics across multiple datasets and random seeds.
- Biological Validation
- Assess whether stability-aware representations preserve biologically meaningful spatial patterns.
Project Deliverables
- StaR Framework Implementation
- An open-source Python implementation compatible with common spatial transcriptomics toolchains.
- Stability Benchmarks
- Comprehensive evaluations demonstrating reduced performance variance across seeds.
- Visualization Tools
- Tools for visualizing performance landscapes, stability surfaces, and spatial domain consistency.
- Documentation and Tutorials
- Clear examples enabling researchers to adopt StaR in their own analyses.
Impact
StaR addresses a critical yet underexplored challenge in spatial transcriptomics: model instability and poor reproducibility. By shifting the focus from single-run performance to stability-aware representation learning, this project improves the reliability and trustworthiness of spatial domain identification methods. StaR has the potential to become a foundational component in robust spatial transcriptomics pipelines and to inspire broader adoption of stability-aware principles in biological representation learning.